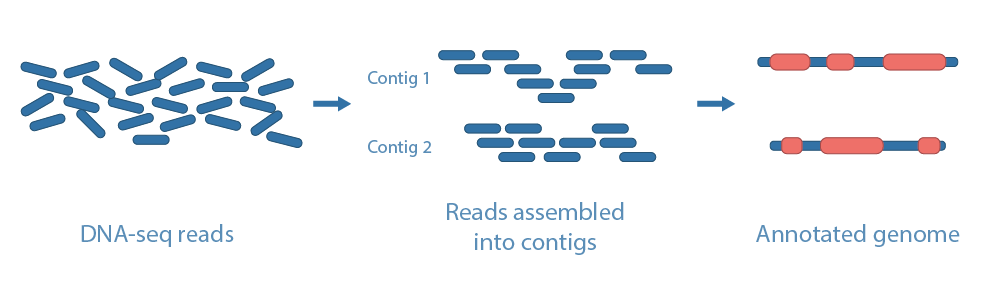
The goal of de novo genome assembly is to determine the sequence of a genome using only randomly sampled sequence fragments. This technique is usually employed for organosms with no reference genome or highly dynamic genomes. In de novo assembly, random fragments from a genome are sequenced and computationally stitched together to generate sequence contigs and scaffolds. The key aspects for achieving high quality de novo assemblies, particularly on highly repetitive genomes, are read length and the assembly algorithm. An assembled genome can then be annotated based on sequence homology, predicted gene sequences and, if available, RNA-sequencing data from the same organism. If annotated genomes for close relative species exist, the annotation can be improved by transferring gene information to the newly assembled genome. The quality of an assembled genome is assessed using metrics such as N50, L50 and completness with regards to highly conserved orthologs.
Due to the high variablility between de novo assemblers, a conjugation of the following workflows is recommended in order to optimize contig length (e.g. N50), genome coverage and maximum, median and average contig size.
Repeat identification and masking is usually a previous step to the gene prediction and annotation phase. The term 'masking' means transforming every nucleotide identified as a repeat to an 'N', 'X' or to a lower case a, t, g, or c (the latter is known as soft masking). The masking step signals to downstream sequence alignment and gene prediction tools that these regions are repeats. Identifying repeats is complicated by the fact that repeats are often poorly conserved; thus, accurate repeat detection usually requires a repeat library for the species of interest.
A good metric of assembly quality is evaluating the completeness. This consists of assessing the extent to which the assembly recovers single-copy orthologs that are present across higher taxonomic groups. The expectations for these genes to be found in a genome/transcriptome in single-copy are evolutionarily strong. If they cannot be identified, it is possible that the sequencing and/or assembly have failed to capture the complete expected gene content.
Annotation of the assembly genome can be divided into structural annotation and functional annotation. In structural annotation, gene prediction tools are used to locate genes within the scaffolds, and the structures of introns, exons, and untranslated regions (UTRs) constituting these genes. In the functional annotation step, homology search and ontology mapping are performed using structure-annotated sequences in order to determine the functions of the genes.