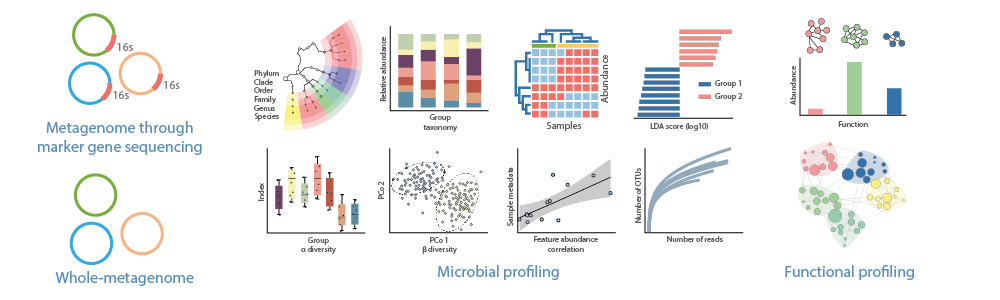
Metataxonomics provide a cost-effective means to identify a wide range of organisms since they rely on rRNA gene sequences for taxonomic profiling. These rRNA markers include 16S rRNA for bacteria, 18S rRNA for eukaryotes, and the internal transcribed spacer (ITS) regions of the fungal ribosome for fungi. These markers work well for phylogenetic profiling because they are ubiquitously present in the population, they have hypervariable regions that differentiate species and they are flanked by conserved regions that can be targeted by "universal" primers. In the analysis of these sequences, two major approaches for representative sequence selections can be employed, clustering to operational taxonomic units (OTUs) and denoising to amplicon sequence variants (ASVs). ASVs have been proposed to replace the limitations in OTU clustering.
In this first stage, raw reads will be demultiplexed (i.e assigning barcoded reads to the respective sample) and a quality filter will be imposed. The quality filtering parameters impose limits on the: (i) minimum number of consecutive high-quality base calls; (ii) maximum number of consecutive low-quality base calls; (iii) maximum number of ambigous (N) characters allowed in a sequence and (iv) the minimum allowed base quality score (Phred quality score). At last paired-end reads, when applicable) are merged to obtain amplicon sequences, and barcodes and primers are removed.
Picking representative sequences as proxies for species is a key step of these types of amplicon analysis. Two major representative approaches are available, OTU clustering and ASV denoising.
A feature table is obtained quatifying the frequency of the feature sequences in each sample. To this feature table, a taxonomical assigment will be performed (kingdom, phylum, class, order, family, genus and species).
At this stage, alpha and beta diversity will be evalutated to assess microbiota diversity. A phylogenetic tree will be generated based on the detect species. Co-occurence, correlation and discrimination of phylotypes by condition or group will also be carried, if applicable.
In this last step, metabolic predictions and functional annotations for the collections of phylotypes will be evaluated.