Whole-metagenome can reach higher resolutions of taxonomical annotation and provide functional gene profiles directly, comparatively with marker gene sequencing. Additionaly, this strategy alleviates biases from primer choice and enables the detection of organisms across all domains of life. The disadvantage of this approach is the cost. Additionally, collections involving a host species should be carefuly considered. Host's DNA can easily be present in higher proportions comparatively with the microbial DNA, which will translate in a lower number of microbial reads which can impair results. Metagenomic classification can be performed by matching reads against a database or assembling reads into contigs, followed by contig match to a database to identify the taxon of each sequence.
In a whole-metagenome analysis the first step is the quality control of the reads by trimming and removing of low quality bases/reads. After this quality control step, in metagenomic experimental steps involving a host species, host reads need to be removed prior to the following analysis. For enviromental samples, this step does not need to be performed.
After quality control, the reads can either be assembled into longer contiguous sequences (assembled into contigs) or passed directly to the taxonomic classifier. The latter is useful for quantitative community profiling and identification of organisms with close relatives on curated databases. When no close relative exist, as can occur in environmental samples, assembly of reads into contigs can be useful since they allow a better assessement of single-copy and conserved genes, taxonomic classification, genome completeness.
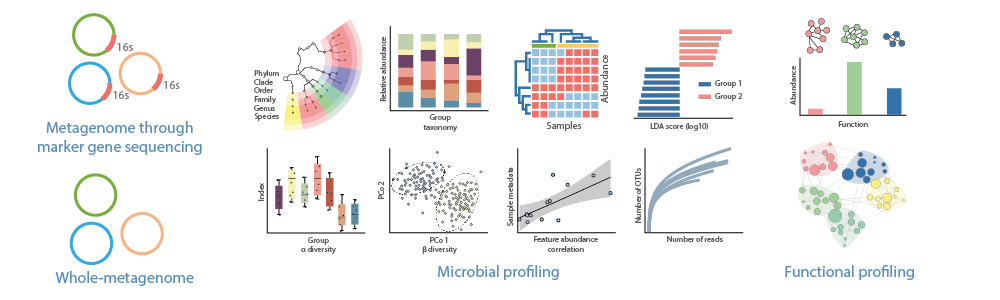
A feature table is obtained quatifying the frequency of the feature sequences in each sample. To this feature table, a taxonomical assigment will be performed (kingdom, phylum, class, order, family, genus and species).
At this stage, alpha and beta diversity will be evalutated to assess microbiota diversity. A phylogenetic tree will be generated based on the detect species. Co-occurence, correlation and discrimination of phylotypes by condition or group will also be carried, if applicable.
Functional annotation is performed using the genes present in the dataset which are combined into modules and pathways for more comprehensive information.