In general, the goal of the majority of DNA sequencing projects is to discover genomic differences (variants) either between healthy and diseased tissues, between individuals of a population, or between strains of an organism. These variations can provide mechanistic insight into disease processes and the function of affected genes. These variants can take the form of single nucleotide variants (SNVs), small DNA insertions or deletions (indels), copy number variations (CNVs), or other structural variants (SVs). Identification of these variants is usually achieved by resequencing strategies where variations are identified by the precise alignment of sequenced reads to a reference genome. Resequencing strategies can range from whole-genome, whole-exome and target sequencing. With the exception of array genotyping, all next generation sequences follow the broad workflow description stated below.
Raw sequence data obtained from a sequencing service provider, in general, is not immediately ready for variant discovery. A first quality control (QC) step is recommended for the assessement of contaminations, bias and errors in raw sequences. By addressing these issues we can reduce the impact of these errors in downstream analysis.
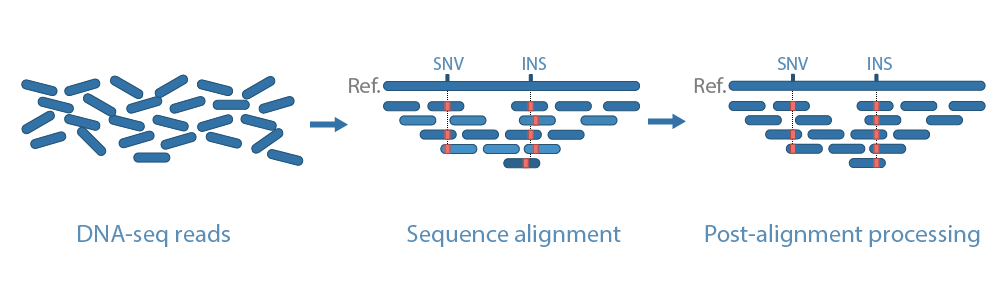
In this step, reads are aligned to a reference genome to determine its precise location.
After the alignment step, technical biases from the underlying sequencing platform and or sample preparation steps can still linger which affect variant calling. Two of the most common sources of biases are duplicates (PCR duplicates and optical duplicates) and uneven base quality scores between sequencing cycles. By identifying duplicates and recalibrating base quality scores we can reduce the impact of these sources of bias from the analysis. Base quality score recalibration can only be performed in organisms with public variation data (VCF) which typically are model-organisms. For the remaining organisms, this step will not be performed.