Copy number variants (CNVs) are regions of the genome containing additional DNA copies (duplications or amplifications) or losses of genetic material (deletions). These variants have been identified across all domains of life, from bacteria and archaea to plants and animals and are and important source of genetic diversity. Although CNVs are a subtype of structural genome variants, their definition and criteria are still evolving. Currently, the size of CNVs is defined from 50bp to several MB.
At this stage, identification of copy-numbers can be assertained using the post-alignment processed file. CNV detection methods can be categorized into five different strategies, including: (1) paired-end mapping (PEM), (2) split read (SR), (3) read depth (RD), (4) de novo assembly of a genome (AS). Each of these approaches has strenghts and limitations depending on the size of CNV and genome coverage. For this reason, a combinatorial approach of the different algorithms is typically recommended.
After CNV discovery filtration is recommended to reduce the numer of false positives resulting from experimental biases. Two such examples of biases are the GC content and the presence of repetitive DNA elements. The first can lead to certain genomic regions being over or under-sampled. The latter can result in large differences between the number of unambiguously mapped reads to a region the number of reads sequenced from that region.
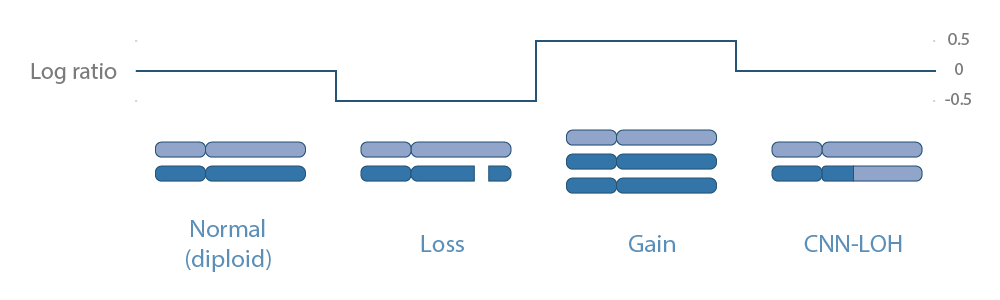
At this stage, filtered CNVs, typically defined as: delection, loss, normal, gain or amplification will be annotated regarding their gene(s)/transcript(s).