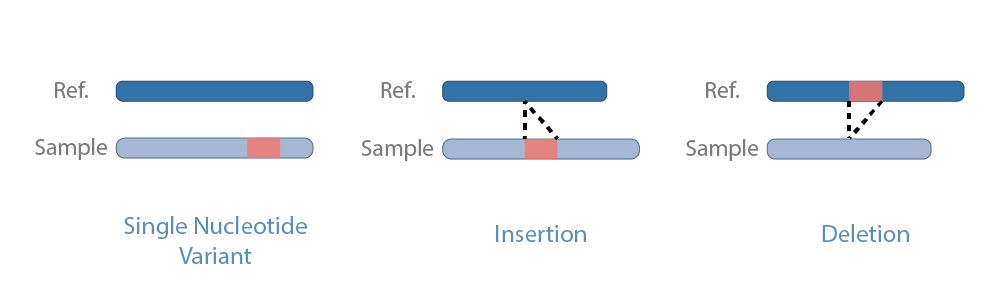
Single-nucleotide polymorphisms (SNPs) and small insertions/deletions (indels) are the two most common types of genetic variants found in genomes.
Variant calling is a method of identification of the differences between the aligned sequenced reads and the reference genome. A wide range of variant callers can be found in the literature aimming to mask alignment and sequencing artifacts, two of the main issues driving the missidentification of variants.
After the variant calling stage, raw SNV and indels in the Variant Call Format (VCF) are obtained. At this stage it is usually recommended either setting hard filters on the data or performing other more complex methodologies (e.g. GATK's Variant Quality Score Recalibration).
At last, the set of filtered variants are functionaly annotated regarding their gene(s)/transcript(s), assessing their influence on protein sequence, and equating the variant with known genomic annotations. Using bioinformatic tools, the seriousness and impact of variants will be quantified.