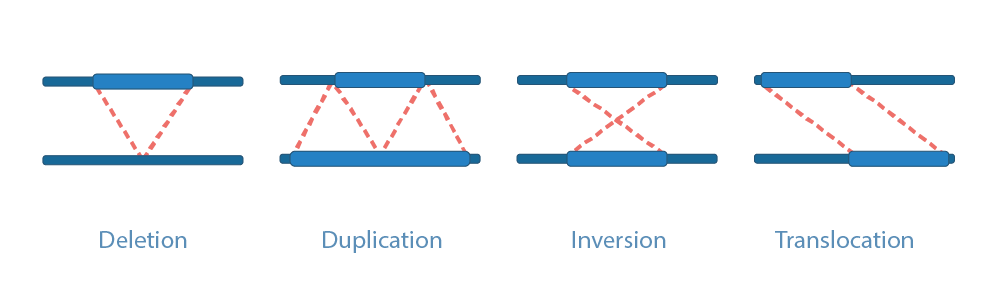
Structural variations (SVs) are an extremly diverse category of genetic variations. SVs comprise a myriad of subclasses, from CNVs to rearrangements (inversions and interchromosomal and intrachromosomal translocations). Additionaly, they include mobile element insertions, sergmental duplications and complex rearrangements containing multiple combinations of the above events. Although short-read sequency can resolve SNVs, this sequencing strategy has difficulties in resolving all above SVs structures given the limited sequence and insert size.
SV detection algorithms leverage signatures that result from mapping discordance between a sample read and the reference genome. Although different heuristics exist, and ensemble approach comprised by containing two or more heuristics has been shown to increase concordance of SV calls when compared with reference data sets.
Following detection, the resultant calls are merged, combining SVs calls uniquely made by each algorithm.
At this stage, filtered SVs, will be annotated regarding their gene(s)/transcript(s).