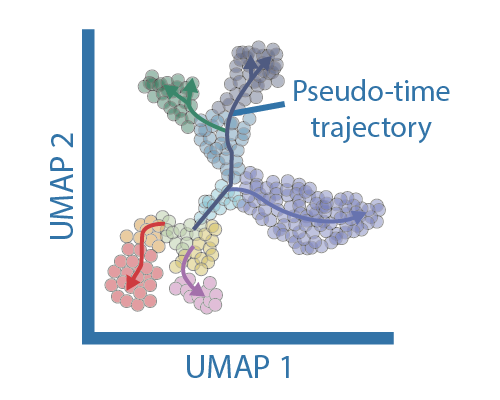
Traditional clustering methods that assign cells to a group are not the most appropriate tools for datasets representing developemental processes or derived from a time-course experiments, where is more appropriate to view cells as a continuum. Continous trajectory analysis are often refered as "pseudotime" and require external information (e.g. sampling time from a time-course experiment or marker genes for developmental trajectories) since they cannot determine the direction or speed of cells allong the trajectory. Most tools for trajectory analysis take one of two approaches: 1) a low-dimensional manifold or 2) unsupervised clustering. Cluster-based pseudotime methods tend to be more accurate for the cases where there is an unequal density of cells (i.e. more cells from one state than the other) and large scale developmental hierarchies. Manifold approaches work best under even sampling across transitions and when examining details of singular transitions. Newer methodolies can infer time dynamics based on the relative abundaces of exonic and intronic reads, which enable the inference of the direction and estimate the rate of chance that each cell is moving in the expression space. This approach is however limited by sequencing depth and the number of reads mapped to introns.